Note
Click here to download the full example code
Guassian Sims¶
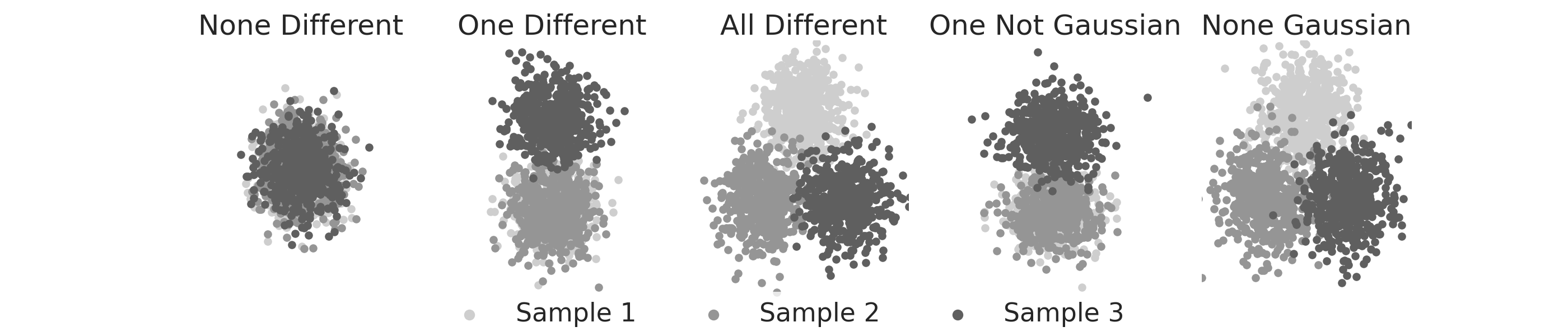
Gaussian k-sample simulations are found in hyppo.tools. Here, we visualize
what these
simulations look like. We use these Gaussian simulations when comparing our
algorithms against multivariate analysis of variance (MANOVA).
import matplotlib.pyplot as plt
import seaborn as sns
from hyppo.tools import gaussian_3samp
# make plots look pretty
sns.set(color_codes=True, style="white", context="talk", font_scale=2)
PALETTE = sns.color_palette("Greys", n_colors=9)
sns.set_palette(PALETTE[2::2])
# constants
N = 500
CASES = [1, 2, 3, 4, 5]
# make a function to plot the Gaussian simulations
def plot_gaussian_sims():
"""Plot simulations"""
fig, ax = plt.subplots(nrows=1, ncols=5, figsize=(28, 6))
sim_titles = [
"None Different",
"One Different",
"All Different",
"One Not Gaussian",
"None Gaussian",
]
# plt.suptitle("Gaussian Simulations", y=0.93, va="baseline")
for i, col in enumerate(ax):
sim_title = sim_titles[i]
# rotated k-sample simulation
sims = gaussian_3samp(N, epsilon=4, weight=0.9, case=CASES[i])
# plot the nose and noise-free sims
for index in range(len(sims)):
col.scatter(
sims[index][:, 0],
sims[index][:, 1],
label="Sample {}".format(index + 1),
)
# make the plot look pretty
col.set_title("{}".format(sim_title))
col.set_xticks([])
col.set_yticks([])
col.set_xlim(-5, 5)
if CASES[i] not in [2, 4]:
col.set_ylim(-5, 5)
sns.despine(left=True, bottom=True, right=True)
leg = plt.legend(
bbox_to_anchor=(0.5, 0.17),
bbox_transform=plt.gcf().transFigure,
ncol=3,
loc="upper center",
)
leg.get_frame().set_linewidth(0.0)
for legobj in leg.legend_handles:
legobj.set_linewidth(5.0)
plt.subplots_adjust(hspace=0.75)
# run the created function for the Gaussian simulations
plot_gaussian_sims()
Total running time of the script: ( 0 minutes 0.217 seconds)